The genetic control of immunoglobulin production solves with elegance and creativity what could be a serious problem.
|
|
|
The variable and constant domains of immunoglobulins are coded by different genes, but not in sufficient numbers for the enormous diversity of immunoglobulins that are produced by an immune response. |
In order to resolve this apparent limitation, the production of immunoglobulins has a special genome organization and various expression mechanisms, permitting great diversity with fewer fixed genes. |
|
|
| THE DIVERSITY OF ANTIBODIES can be ensured thanks to: |
| The existence of multiple fixed genes V, D and J in the stem cell. |
| Modification of DNA when developing B cells become mature B cells. |
| Multiple possibilities of recombination of heavy chains with light chains. |
| Somatic mutations. |
| Multiple recombinations. |
|
Genetic organization of porcine immunoglobulins.
The genetic organization of immunoglobulin formation has been studied in depth in humans and murine species, with the porcine species becoming increasingly better known.
Compared to most proteins which are coded by single genes (maternal and paternal copies), immunoglobulins have multiple versions of genes. Each B lymphocyte precursor can choose between any of the different versions available that code the different regions. This phenomenon, which is totally random, is the main basis enabling the immune system to produce highly diverse antibodies. In conclusion, genes can be reorganised randomly to synthesize antibodies, and the B lymphocyte precursors can produce hypermutation processes. Combined with multiple combinations between heavy and light chains, this enables a huge diversity of antibodies induced in the immune response to be expressed or coded with relatively few fixed genes. |
|
| Immunoglobulin genes
There are three groups of genes linked to immunoglobulin coding, located in different chromosomes.
The genes that code the constant and variable regions of immunoglobulins are separated within the genome of all the cells of an organism, except in B lymphocytes. These genes become closer during the maturing process of the B cells, enabling random processes of somatic recombination to take place and the possibilities of diversity to be increased.
These genes consist of three different loci, known as:
| H Locus. Code: Heavy chains, variable and constant domains. |
Variable domain |
V |
| D |
| J |
| Constant domain |
Cm |
| Ca |
| Cg |
| Ce |
| Kappa Locus (K). code: Light chains K |
V |
| J |
| Lambda Locus (l). Code: Light chains l |
V |
| J |
|
The H locus codes the heavy chains of immunoglobulins. It consists of two gene groups. One group codes the variable domains of heavy chains and consists of three different genes:
|
|
V genes. So named as they code variability |
|
D genes. Code diversity. |
|
J genes. Joining. |
|
|
| The V genes, which are the most abundant, code most variable domains. Only one family of this gene has been identified in porcine species.
The D genes are very diverse with between 1 and more than 15 segments having been identified. They are very variable, with the greatest number of variations in the nucleotide sequence of the three types of H locus genes.
The J genes code the variable region. Only one J segment has been found in pigs (there are four in mice and nine in humans). |
The constant part of the heavy chains is coded by the genes Cm, Ce, and Ca. Only one type of gene has been found in pigs. These genes code the heavy chains of IgM, IgE and IgA immunoglobulins, respectively. However, eight copies of genes have been described corresponding to gene Cg. The study of these genes has proved that there are at least five subclasses of IgG in pigs. These are coded by genes Cg1, Cg2a, Cg2b, Cg3 and Cg4. |
The kappa locus (K) only has one segment, known as CK. The lambda locus (l) consists of two genes and two segments. The genes located on these loci are responsible for coding the kappa and lambda light chains. |
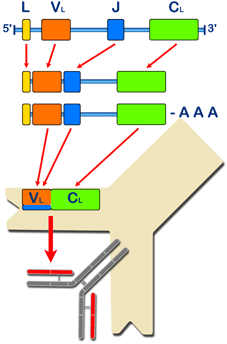
Diagram of the production of the light chain of porcine immunoglobulins. |
|
The change of isotype of the immunoglobulins is also genetically controlled. The immune response generally starts with the production of IgM and then IgG or IgA or IgE, whose Fc fragment has different biological properties. On the 5' side of all CH genes, there is a sequence named S that enables a change in isotype. Differences are produced by gene deletion. |
|
Known immunoglobulin variants.
Monoclonal antibodies have enabled the genetic variability of porcine immunoglobulins to be studied. Three types have been discovered: |
| ISOTYPIC Variants |
| ALLOTYPIC Variants |
| IDIOTYPIC Variants |
|
|
|
The isotypic variants are coded by genes which are found in the same animal species, meaning that this type of immunoglobulin is common amongst all members of the porcine species. The part of the immunoglobulin molecule which allows different isotypes to be differentiated is found in the constant regions of the heavy chains. Four main immunoglobulin isotypes have been classified in pigs. These are called IgM, IgA, IgG e IgE. |
Allotypic variants are only found in the immunoglobulins of some animals of the porcine species. They are allelic markers which are expressed in the constant regions of heavy and light chains of the immunoglobulins of some animals of the same species.
|
|
Idiotypic variants can be found in a single individual of the same species or in several, depending on whether they are in contact or not with the same antigen. They are induced by the different antigen epitopes and are found in the hypervariable regions of heavy and light chains. |
|

